|
Andrographis paniculata Transcriptome Analysis Summary |
|
|
|
Raw Data used for the genic-SSR mining and primer
designing
|
|
De novo meta-transcriptome assembly using our in-house RNA-Seq data of
A. paniculata generated through
Roche-454 GS-FLX platform (SRA Acc. No. SRR719255), and other publically
available RNA-Seq data of the same species and tissue generated through
different sequencing platforms including Illumina (SRA Acc. No. SRR1519324,
SRR12791806, SRR12791807, SRR1292497), and semiconductor-based technology Ion
Torrent
(SRA Acc. No. SRR8500525) generates a total of 98,514 non-redundant transcripts.
These 98,514 non redundant transcripts further used for the identification of
microsatellite markers, their primer pairs, and transcription factors.
|
|
Identification of Microsatellite markers (SSRs)
Microsatellite marker (SSR) identification has been performed using Krait
v1.3.3 software with the parameters: six repeat units for di-nucleotide, and
five repeat units for tri-, tetra-, penta-, and hexa-nucleotide repeats and
number of bases interrupting two SSRs in a compound microsatellite to be 100 bp.
Table 1. Summary statistics of the perfect SSRs predicted using
the Krait v1.3.3 software.
|
|
Item
|
Description
|
Number
|
|
Number of perfect SSRs
|
Counts
|
37,814
|
|
Total length of perfect SSRs
|
Bp
|
651,825
|
|
Average length of perfect SSRs
|
Total length (bp)/No. of SSRs
|
17.24
|
|
Number of compound SSRs
|
Counts
|
1,462
|
|
Number of imperfect SSRs
|
Counts
|
170,017
|
|
Total length of imperfect SSRs
|
Bp
|
4,825,112
|
|
|
|
|
Table 2. Distribution of mono- to hexa- type of perfect SSRs.
|
|
SSR Type
|
No. of SSRs
|
Length (bp)
|
Percent (%)
|
Average
length (bp)
|
Relative
abundance (loci/Mb)
|
Relative density (bp/Mb)
|
|
Mono
|
5,554
|
87,503
|
14.69
|
15.75
|
48.81
|
769.02
|
|
Di
|
17,301
|
286,566
|
45.75
|
16.56
|
152.05
|
2518.48
|
|
Tri
|
12,534
|
221,274
|
33.15
|
17.65
|
110.15
|
1944.66
|
|
Tetra
|
1,781
|
38,176
|
4.71
|
21.44
|
15.65
|
335.51
|
|
Penta
|
425
|
11,160
|
1.12
|
26.26
|
3.74
|
98.08
|
|
Hexa
|
219
|
7,146
|
0.58
|
32.63
|
1.92
|
62.8
|
|
|
|
|
Table 3. Distribution of di- to hexa- type of perfect SSRs with
three pair of primer.
|
|
SSR Type
|
No. of SSRs
|
Length (bp)
|
Percent (%)
|
Average
length (bp)
|
Relative
abundance (loci/Mb)
|
Relative density (bp/Mb)
|
|
Mono
|
2,547
|
36,108
|
11.26
|
14.18
|
22.39
|
317.35
|
|
Di
|
9,149
|
144,542
|
40.45
|
15.80
|
80.41
|
1270.36
|
|
Tri
|
9,351
|
160,002
|
41.34
|
17.11
|
82.18
|
1406.24
|
|
Tetra
|
1168
|
24,684
|
5.16
|
21.13
|
10.27
|
216.94
|
|
Penta
|
277
|
7,255
|
1.22
|
26.19
|
2.43
|
63.76
|
|
Hexa
|
128
|
4,194
|
0.57
|
32.77
|
1.12
|
36.86
|
|
|
|
 |
|
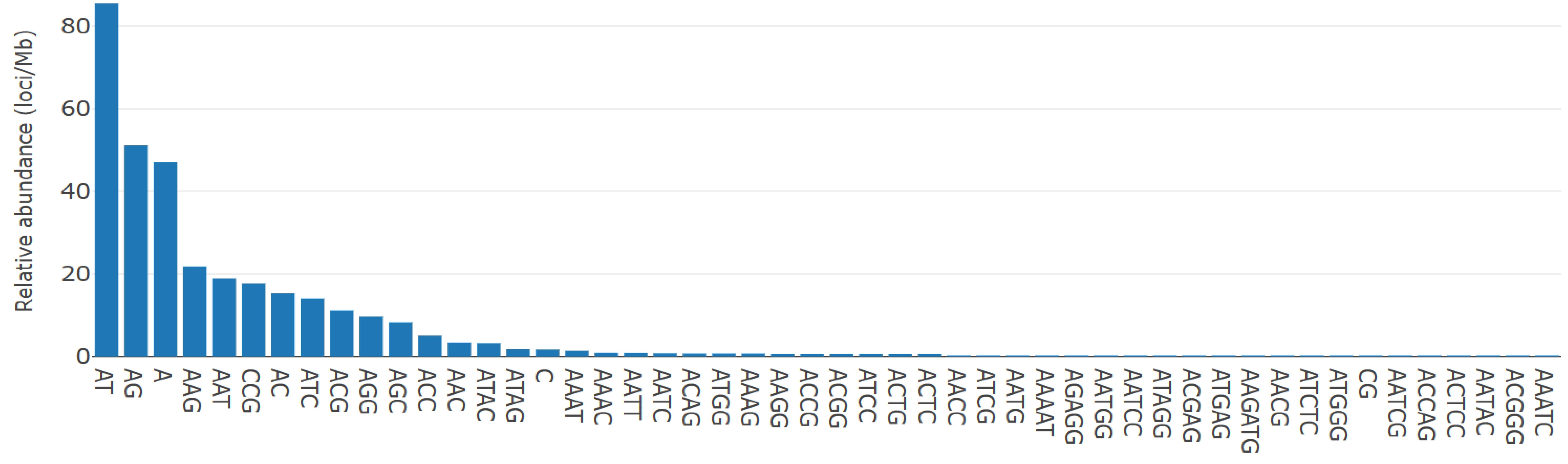
Figure 1. Distribution of most abundant type of motif
categories.
|
 |
|
Figure 2. SSR repeats distribution of each mono to hexa SSR
types.
|
|
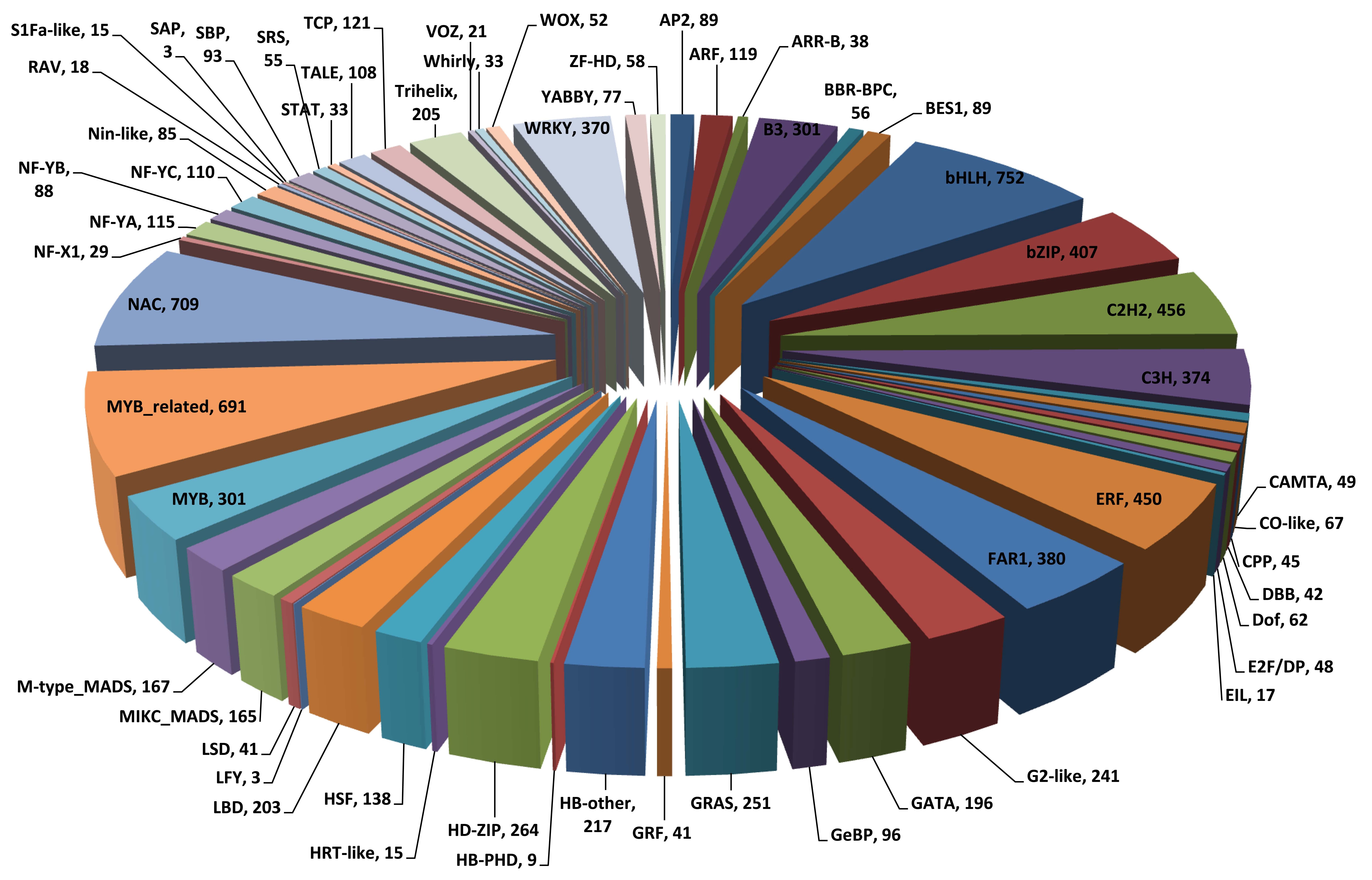
Figure 3. Distribution of different transcription factor
categories predicted in A. paniculata trancriptome.
Table 4. Gene families related to biochemical compound
biosynthesis present in A. paniculata.
|
S. No.
|
Family Name
|
No. of Transcripts
|
Function |
|
1.
|
Cytochrome P450
|
359
|
Biosynthesis of secondary metabolites |
|
2.
|
Protein Kinases
|
2,762
|
Signal transduction pathways |
|
3.
|
Heat Shock Proteins (HSPs)
|
70
|
Therapeutic treatments |
|
4.
|
Transporters
|
1,593
|
Transport of metabolites |
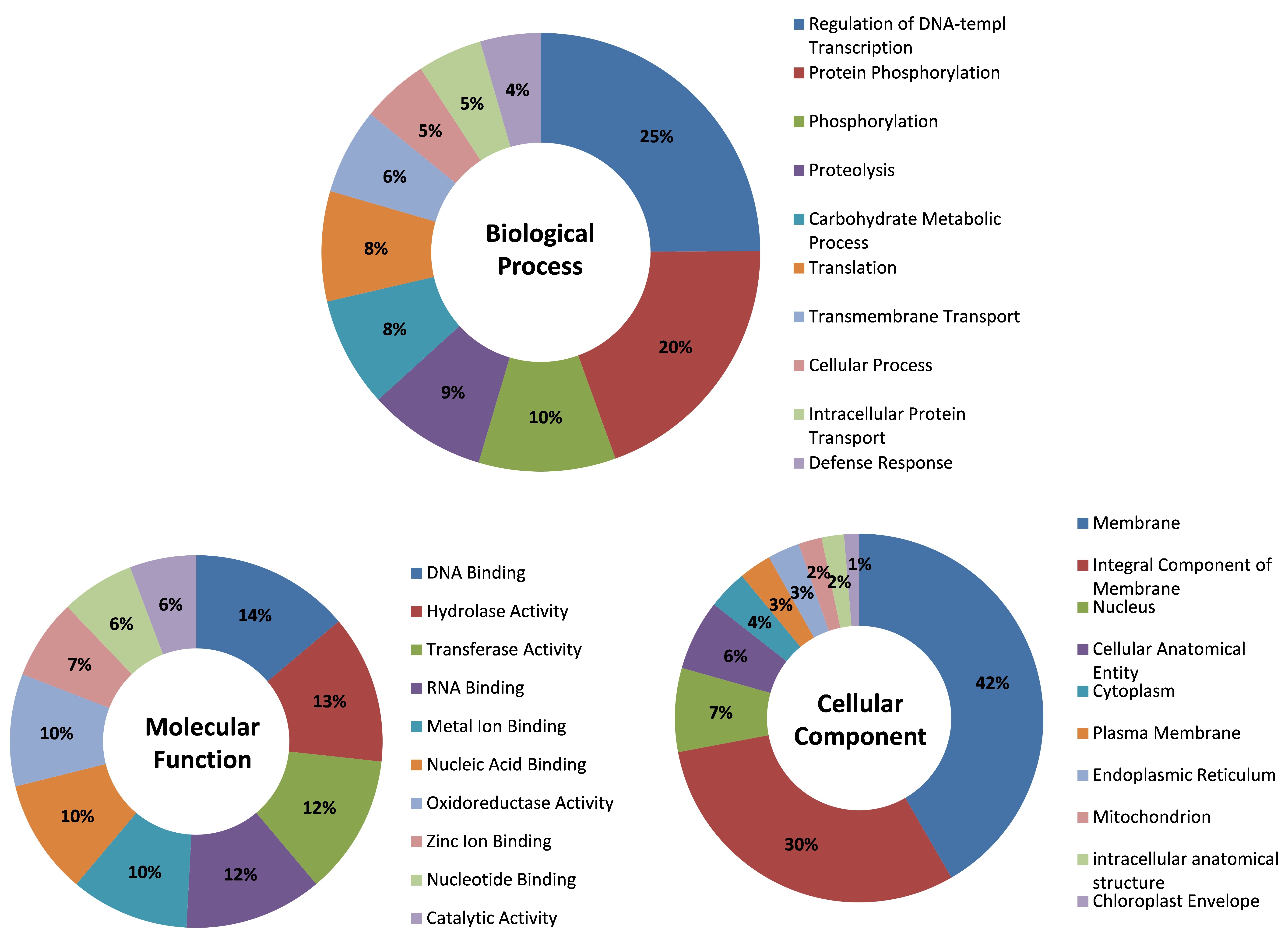
Figure 4. Distribution of functionally annotated transcripts
of A. paniculata in three main categories of GO classification
viz. Biological Process, Molecular Function, and Cellular Component.
|
|
|
| Copyright ©
Reg. No. SW-15900/2022 |
 |
ICAR- National Bureau of Plant Genetic Resources, New Delhi, India |
|
| This work is supported by
Indian Council for Agricultural Research, Government of India, New Delhi |
|