|
Expand All
|
|
|
- To manage and promote sustainable use of plant genetic and genomic resources of agri-horticultural crops and carry out related research
- Molecular profiling of varieties of agri-horticultural crops and GM detection technology research
The National Research Centre on DNA Fingerprinting was established by the Indian Council of Agricultural Research during December 1995 in ICAR-National Bureau of Plant Genetic Resources (NBPGR) with the objective to develop and standardize molecular marker techniques for identification of agricultural crop varieties, important genetic stocks, landraces and other economically important plant genetic resources of the country. The National Research Centre on DNA Fingerprinting was renamed as the Division of Genomic Resources in February 2012. Since then, the division has been involved activities, generation, utilization and conservation of plant genomic resources.
|
|
|
Generating and Conserving Genomic Resources
- Genomic resources in the form of 68,925 SNPs were developed using genotyping by sequencing (GBS) of 131 linseed germplasm accessions conserved in the Indian National Genbank (INGB).
- Quantitative trait nucleotides (QTNs) were identified using Genome-Wide Association Study (GWAS) for flowering time (53 QTNs), days to maturity (30 QTNs), plant height (27 QTNs), and thousand seed weight (30 QTNs) in linseed accounting for 35.28%, 28.78%, 36.6%, and 38.65% trait variation, respectively.
- Developed 311 accession of core representing genetic diversity of 3115 accession using SSR based genotyping in safflower germplasm for effective utilization.
- Molecular systematics and species delineation of important vegetable species viz. Luffasps. Cucumissps, Trichosanthescucumerina complex, Alliumsps, Musasps, and Vegetable Amaranthusing DNA based barcoding method.
- Described new taxa Alliumnegianum, Cucumismelo var. alwarensis supported with DNA based barcoding
- Assessment of nuclear DNA content and ploidy analysis of invitro gene bank accessions of cultivated banana and wild species of Indian origin.
- Chromosome identification and localization of rDNA for characterisation of wild species for genomic constitution of Vignasps, Musasps, Luffasps. using Fluroscense in situ hybridization.
- Identification and localisation of alien gene introgression from Aegilopsmarkgrafii in wheat chromosome using Genomic in situ hybridization (GISH).
- A novel gene targeted marker technique CBDP (CAAT Box-Derived Polymorphism) has been developed that can be used for various genotyping applications in plants. The technique exploits conserved CCAAT motif in the CAAT box region of promoter region of plant genes to generate markers. The concept has been validated in three different crops (Jute, cotton, linseed) representing five different species (Corchorus capsularis, C. olitorius, Gossypium hirsutum, Gossypium orboratum) and linseed (Linum usitatissimum).
- A cold tolerance acclimation gene CAS15 was cloned from a cold tolerant ecotype of plants. The gene encodes for a dehydrin like protein and can be exploited for engineering cold tolerance in crops.
- DNA Barcoding loci rbcL, matK, trnH-psbA and ITS region alone and/combination of two loci identified 21 genomic species in Oryza and were used for establishing correct genetic identity of mis-labeled species. Two combined loci DNA barcodes (rbcL + ITS) gave better species delineation and proper barcode gaps for species identification in genus Luffa.
- Molecular Profiling: DNA profiling services were rendered to various public and private sector organizations. The details of the crops and the number of samples DNA profiled are listed in the Table below.
- Black pepper genome sequenced: Under CRP on Genomics, NBPGR has produced 1.2 GB draft sequence of black pepper (Piper nigrum) genome. Large number of genomic resources is expected to be generated.
- Discovery of “Bose anatomy” (non-Kranz C4 photosynthesis) in wheat grains
- Discovery of non-Kranz C4 photosynthesis in two cell layers (cross- and tube-cells) of pericarp in developing wheat grains. Named it as “Bose anatomy” in honor of his earliest works on C4 in Hydrilla reported in 1924 when C3 itself was not known.
Brochure DNA Fingerprinting - MOLECULAR PROFILING OF VARIETIES OF AGRI-HORTICULTURAL CROPS
Brochure - Division of Genomic Resources
DNA Fingerprinting in Plants Standard Operating Methods and Protocols
Brochure - Little millet, A Nutri-cereal:Glimpses of Variability in the National Genebank
SESOMICS March 2022
Number of varieties fingerprinted in recent years
|
Crop
|
Scientific Name
|
2014
|
2015
|
2016
|
2017
|
2018
|
2019
|
2020
|
2021
|
2022
|
Total
|
|
Almond
|
Prunus dulcis
|
-
|
-
|
-
|
4
|
-
|
-
|
-
|
-
|
-
|
04
|
|
Amaranth
|
Amaranthus tricolor
|
-
|
-
|
-
|
-
|
-
|
3
|
-
|
-
|
-
|
03
|
|
Areca nut
|
Areca catechu
|
-
|
18
|
-
|
21
|
-
|
-
|
-
|
-
|
-
|
39
|
|
Bay leaf
|
Laurus nobilis
|
8
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
08
|
|
Barley
|
Hordeum vulgare
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
1
|
01
|
|
Bitter gourd
|
Momordica charantia
|
-
|
-
|
-
|
-
|
1
|
-
|
-
|
-
|
-
|
01
|
|
Brinjal
|
Solanum melongena
|
-
|
-
|
-
|
-
|
-
|
9
|
4
|
-
|
01
|
14
|
|
Buckwheat
|
Fagopyrum sp.
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
1
|
-
|
01
|
|
Chenopodium
|
Chenopodium album
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
1
|
-
|
01
|
|
Chickpea
|
Cicer arietinum
|
3
|
5
|
2
|
4
|
-
|
-
|
-
|
-
|
2
|
16
|
|
Chilli
|
Capsicum annuum
|
-
|
-
|
-
|
-
|
-
|
2
|
2
|
1
|
-
|
05
|
|
Cluster bean
|
Cyamopsis tetragonoloba
|
-
|
-
|
-
|
1
|
-
|
-
|
-
|
-
|
-
|
01
|
|
Cotton
|
Gossypium sp.
|
28
|
37
|
20
|
30
|
17
|
21
|
25
|
40
|
17
|
235
|
|
Cowpea
|
Vigna unguiculata
|
-
|
-
|
-
|
-
|
3
|
2
|
3
|
-
|
-
|
08
|
|
Cucumber
|
Cucumis sativus
|
3
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
03
|
|
Date Palm
|
Phoenix dactylifera
|
-
|
-
|
4
|
-
|
-
|
-
|
-
|
-
|
-
|
04
|
|
Dolichos
|
Dolichos sp.
|
-
|
-
|
-
|
-
|
-
|
-
|
1
|
-
|
-
|
01
|
|
Fennel
|
Foeniculum vulgare
|
-
|
-
|
-
|
-
|
-
|
-
|
2
|
-
|
-
|
02
|
|
Fenugreek
|
Trigonella foenum-graecum
|
-
|
-
|
-
|
-
|
-
|
-
|
1
|
-
|
-
|
01
|
|
Finger millet
|
Eleusina coracana
|
-
|
-
|
-
|
-
|
-
|
2
|
-
|
-
|
-
|
02
|
|
Foxtail millet
|
Setaria italica
|
-
|
-
|
-
|
-
|
-
|
1
|
-
|
-
|
-
|
01
|
|
French bean
|
Phaseolus vulgaris
|
-
|
-
|
-
|
14
|
-
|
-
|
1
|
-
|
-
|
15
|
|
Garlic
|
Allium sativum
|
-
|
-
|
2
|
-
|
-
|
-
|
-
|
-
|
-
|
02
|
|
Garcinia
|
Garcinia sp.
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
3
|
-
|
03
|
|
Horse gram
|
Macrotyloma uniflorum
|
-
|
3
|
-
|
-
|
-
|
-
|
-
|
4
|
-
|
07
|
|
Lentil
|
Lens culinaris
|
-
|
-
|
2
|
1
|
-
|
-
|
-
|
-
|
-
|
03
|
|
Linseed
|
Linum usitatissimum
|
2
|
-
|
-
|
2
|
1
|
-
|
-
|
1
|
1
|
07
|
|
Little millet
|
Panicum sumatrense
|
-
|
-
|
-
|
-
|
-
|
2
|
-
|
-
|
-
|
02
|
|
Maize
|
Zea mays
|
23
|
-
|
5
|
40
|
62
|
10
|
3
|
-
|
3
|
146
|
|
Melon
|
Cucumis melo
|
-
|
-
|
-
|
-
|
2
|
-
|
-
|
-
|
-
|
02
|
|
Mung bean
|
Vigna radiata
|
-
|
3
|
4
|
5
|
-
|
4
|
5
|
8
|
6
|
35
|
|
Mustard
|
Brassica sp.
|
3
|
2
|
2
|
3
|
3
|
-
|
12
|
2
|
5
|
32
|
|
Okra
|
Abelmoschus esculentus
|
-
|
-
|
-
|
-
|
2
|
-
|
5
|
-
|
-
|
07
|
|
Onion
|
Allium cepa
|
-
|
1
|
-
|
-
|
-
|
-
|
-
|
-
|
-
|
01
|
|
Oats
|
Avena sativa
|
-
|
-
|
-
|
-
|
2
|
-
|
2
|
2
|
1
|
07
|
|
Paddy
|
Oryza sativa
|
14
|
8
|
2
|
40
|
9
|
26
|
23
|
1
|
26
|
149
|
|
Pea
|
Pisum sativum
|
-
|
-
|
2
|
1
|
-
|
-
|
1
|
-
|
4
|
08
|
|
Pearl millet
|
Pennisetum glaucum
|
-
|
18
|
4
|
15
|
7
|
4
|
4
|
-
|
-
|
52
|
|
Pepper
|
Piper nigrum
|
-
|
-
|
-
|
-
|
-
|
-
|
6
|
-
|
-
|
06
|
|
Perilla
|
Perilla frutescens
|
-
|
-
|
1
|
-
|
-
|
-
|
-
|
-
|
-
|
01
|
|
Pigeon pea
|
Cajanus cajan
|
-
|
-
|
-
|
13
|
-
|
4
|
-
|
-
|
13
|
30
|
|
Radish
|
Raphanus sativus
|
-
|
-
|
1
|
-
|
-
|
-
|
-
|
-
|
-
|
01
|
|
Ragi
|
Eleusine coracana
|
-
|
-
|
-
|
48
|
-
|
-
|
-
|
-
|
-
|
48
|
|
Raya
|
Brassica juncea
|
-
|
-
|
-
|
2
|
-
|
-
|
-
|
7
|
-
|
09
|
|
Sesame
|
Sesamum indicum
|
-
|
-
|
-
|
8
|
1
|
6
|
-
|
1
|
1
|
17
|
|
Sorghum
|
Sorghum bicolor
|
4
|
4
|
4
|
4
|
-
|
5
|
-
|
-
|
5
|
26
|
|
Soybean
|
Glycine max
|
-
|
-
|
12
|
-
|
27
|
-
|
4
|
3
|
3
|
49
|
|
Sponge gourd
|
Luffa aegyptiaca
|
-
|
2
|
-
|
-
|
-
|
3
|
-
|
-
|
-
|
05
|
|
Sunflower
|
Helianthus annuus
|
-
|
-
|
-
|
1
|
-
|
-
|
-
|
-
|
1
|
02
|
|
Taramira
|
Eruca sativa
|
-
|
3
|
3
|
-
|
-
|
-
|
-
|
4
|
-
|
10
|
|
Urd bean
|
Vigna mungo
|
-
|
13
|
-
|
3
|
-
|
1
|
3
|
14
|
7
|
41
|
|
Walnut
|
Juglans sp.
|
-
|
-
|
-
|
-
|
-
|
8
|
-
|
2
|
2
|
12
|
|
Wheat
|
Triticum aestivum
|
18
|
-
|
4
|
1
|
4
|
4
|
1
|
1
|
18
|
51
|
|
Total
|
106
|
117
|
74
|
261
|
141
|
117
|
108
|
97
|
116
|
1137
|
- Generation of 56,404 unigenes and 3883 EST-SSR markers in finger millet (Eleusine coracana L. Gaertn.); generation of 25213 unigenes and 1305 EST-SSR markers in little millet (Panicum sumatrense), and generation of 1,94,307 transcripts and 18,204 EST-SSR markers in kodo millet (Paspalum scrobiculatum) and submission of information to NCBI.
- Development of a database for Cucumis melo microsatellite markers: A versatile database for Cucumis melo microsatellite markers has been developed. This database is the first online marker resource developed for musk melon, comprising a wide range of genomic datasets that can assist plant breeders and researchers to select the most appropriate markers for crop improvement. This database can also be applied to other cucurbit crops.
- A genome wide analysis, using Restriction site-Associated DNA Sequencing (RAD-Seq), a next-generation sequencing based scaffolds/contigs sequences data and identified 45,066 perfect microsatellite repeat-motifs spanning ~334 Mb of bottle gourd genome. Of the 45,066 microsatellites, about 105 (~0.2%) markers were successfully validated in five accessions of Lagenaria siceraria. These markers were mounted onto linkage maps for bottle gourd and syntenic relationships with the available cucurbits with a genome sequence were studied.
- A large dataset composed of 58,865 unigenes derived from the sponge gourd transcriptome was assembled. All unigenes ranged from 301 bp to 117.9 Kb, with a average length of 1.2 kb, an N50 equal to 1,907 bp and 45.69 % GC content. Based on these generated sequences, we identified 8,934 putative simple sequence repeats (SSRs) and genes in two ToLCNDV resistant (DSG-6) and susceptible lines (Pusa Sneha) of sponge gourd. These transcriptome sequences and EST-SSR data are of great value for the discovery of novel genes involved in ToLC NDV resistance and for marker-assisted selection in sponge gourd.
- Technology Developed for Identification of SRAP Markers Linked to the Single Dominant Resistance Gene against Tomato Leaf Curl New Delhi Virus in Luffa cylindrica Roem: Two sequence-related amplified polymorphism (SRAP) markers closely-linked to the ToLCNDV-susceptible gene in the susceptible parent and in a susceptible bulk population; and two SRAP markers closely-linked to the resistance gene in the resistant parent and in a resistant bulk population were found. These can be used for large-scale screening of genotypes of L. cylindrica for resistance against ToLCNDV at the seedling stage, and to accelerate the breeding of high yielding, ToLCNDV resistant varieties and hybrids.
- Nucleotide-binding site (NBS) domain of resistance gene candidates (RGCs) were cloned from Tomato Leaf Curl New Delhi Virus (ToLCNDV) resistant Luffa cylindrica (sponge gourd) genotype, DSG-6. Sixteen non-redundant sequences of RGCs were identified with un-interrupted open reading frames (ORFs) and high amino acid sequence homologiesNBS-LRR proteins from GenBank database. The comparative analysis of expression profiles of sgRGCs in asymptomatic and field-driven symptomatic leaf tissues of ToLCNDV resistant and susceptible genotypes revealed RGCLc28 is expressed consistently in resistant genotypes. The differentially expressed RGCLc28 of DSG-6 is predicted to have strong association with the resistance trait against the leaf curl and mosaic disease in sponge gourd and may serve as an important genomic resource for candidate gene discovery.
- Development of a core for North-eastern rice collection (7000 accessions) using Single Nucleotide Polymorphism (SNP) markers.
- Molecular profiling of 800 rice varieties and 200 landraces was completed with SSR/SNP markers and a rice database has been developed.
- Genomic-SSR (g-SSR) markers were generated for Kalmegh (Andrographis paniculata) and Giloe (Tinospora cordifolia) using genomic enrichment method
- Developed EST-SSR and transcription factor database for Kalmegh (Andrographis paniculata) and Giloe (Tinospora cordifolia).
- DNA barcoding of Luffa sps.: Sequences for Luffa (39) accns. representing L. acutangula complex (15 accns.) and L. cylindrica (19 accns.) were generated for barcoding loci— trnHpsbA (non-coding spacer), rpoC1 and ycf5 (coding plastid gene). Out of three loci, rpoC1 and ycf5 did not provide informative sequence sites for differentiation of species. However, trnH-psbA spacer clearly distinguished two species and provided variation within L. acutangula complex where earlier species of L. hermaphrodita is included. This result along with other informative barcoding loci would solve the species status of L. hermaphroidita.
Biosystematics studies
Comprehensive studies on Biosystematics of the genera Vigna, Cucumis and Abelmoschus helped to solve several of the outstanding problems related to species identities and relationships in the above three genera. In addition, the study involved involved established Systematists, Cytogeneticist, Plant Genetic Resources persons and Molecular Taxonomists.
Comprehensive data on occurrence, distribution and prevalence of diversity in species under the genera Vigna, Cucumis and Abelmoschus in India was collected. The study succeeded in generating substantial amount of information on the target genera, in addition to producing over 1500 interspecific derivatives and stabilized lines. Further several valuable genetic resources have been identified as source of genes for tolerance to Mungbean Yellow Mosaic Virus in Vigna, Yellow Vein Mosaic Virus and Enation Leaf Curl Virus in okra, carotenoid rich germplasm in cucumbers, Downey Mildew resistant lines in melons, etc. The species diversity described and assembled includes over 500 accessions belonging to 22 species and 2 varieties of Vigna; 12 species and 5 varieties of Cucumis; and 13 species of Abelmoschus. These along with over 1500 interspecific derivatives covering all species under these three genera form a highly valuable genetic resource for crop improvement programs since these have been shown to contain the valuable genes of economic importance that could not pass through the barriers of domestication bottlenecks during the process of bringing wild species into cultivation.
The studies conducted have effectively resolved the long standing scientific problems related to the following major aspects which had remained unresolved so far:
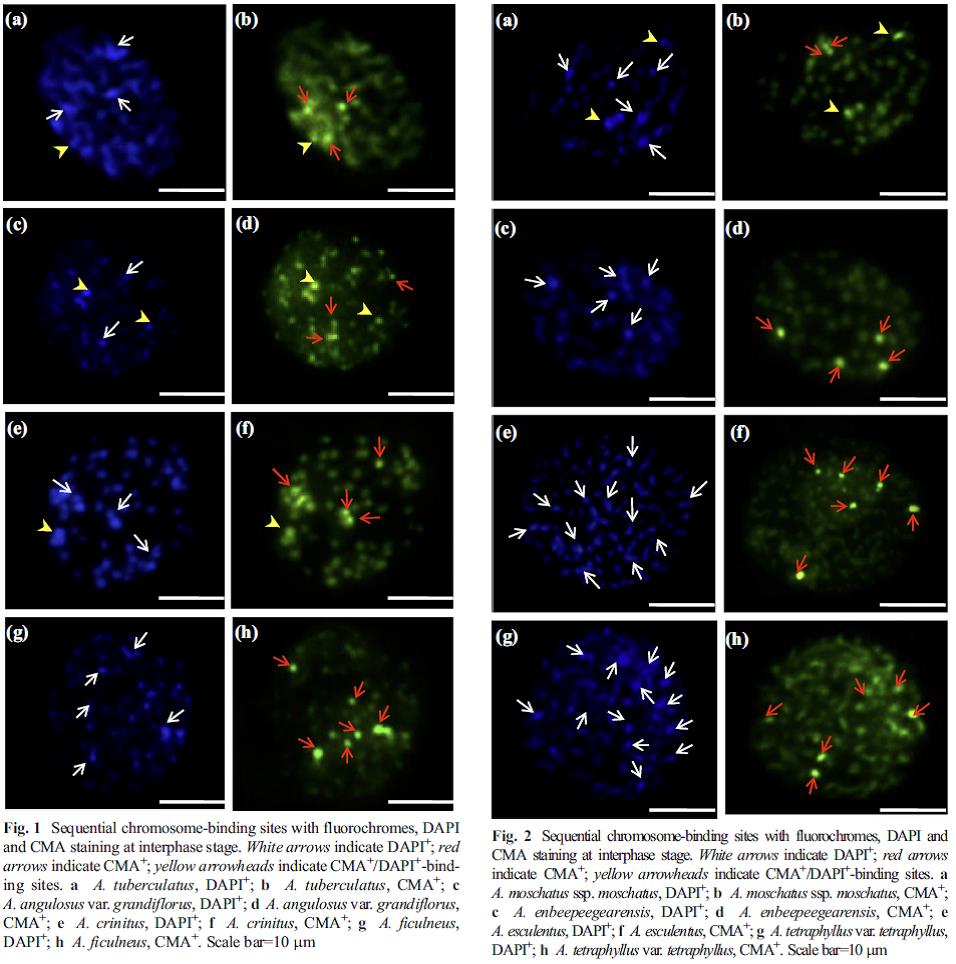 |
| Figure 1. Analyses of sequential chromosome-binding sites with fluorochromes, DAPI and CMA staining at interphase stage in Abelmoschus species indicating species differentiation. |
| |
| |
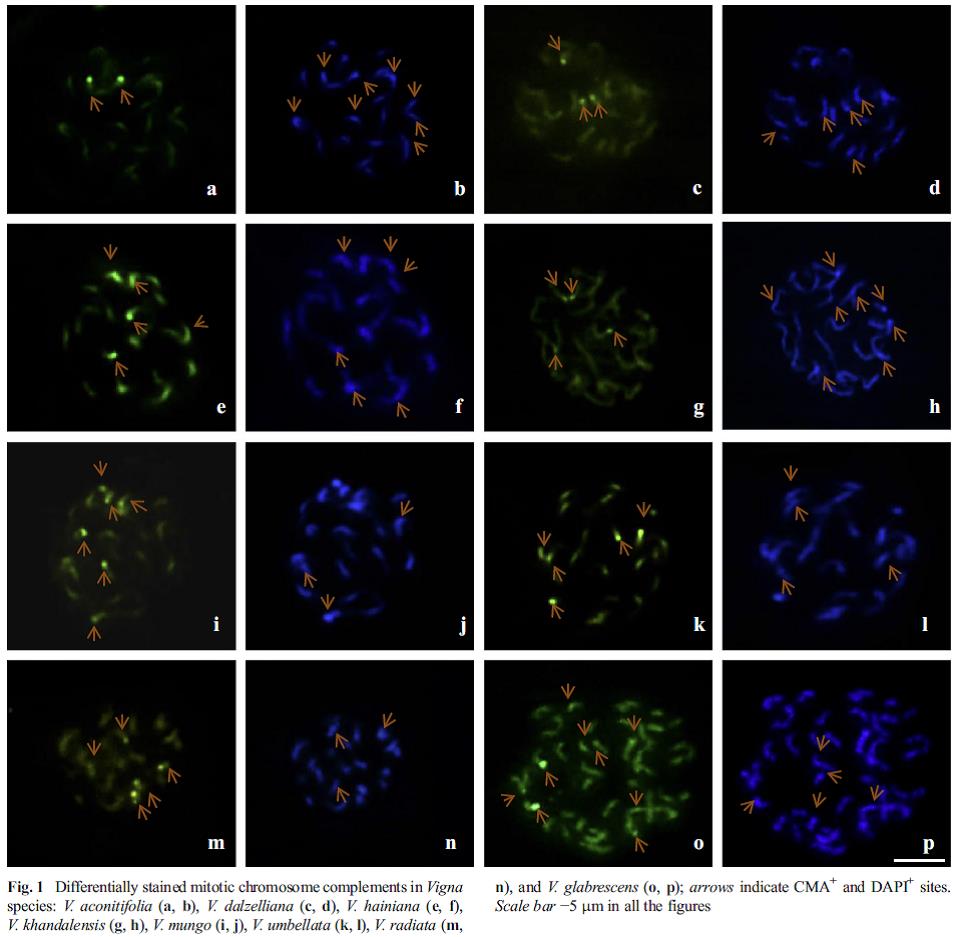 |
| Figure 2. Differential staining of mitotic chromosome complements in 8 Vigna species depicting chromosomal differentiation in the genus. |
- Establishment of identities of the naturally occurring populations and species of the genera Vigna, Cucumis and Abelmoschus in India and describing their identification characteristics thereby refining the taxonomy of the genera Vigna, Cucumis and Abelmoschus.
- Relationships among the species based on macro and micro-morphological variations as well as cyto-morphological features which led to formulation of keys for identification of species on the basis of both morphological features and seed surface micro-morphology.
- Confirmation of the evidences based on molecular cytology and molecular taxonomic studies and to identify the progenitor species of the cultivated Vigna group of pulses, cucumber and okra. This led to devising of methodologies for cytological studies in these difficult genera and molecular cytology using GISH and FISH to describe phylogeny based on chromosomal affinities (figure 1 & 2).
- Understanding the effect of domestication process on genes conferring resistance to yellow mosaic virus in Vigna. The study resulted in identification of accessions of wild Vigna species with tolerance to mungbean yellow mosaic virus which are new sources of genes to combat this devastating virus in mung and urd.
- Transfer of resistance genes from wild species to cultivars genetic background and bring about genetic enhancement of the cultivated gene pool for the target traits was achieved during the course of crossability studies which resulted in generation of colchiploidized allopolyploids in over 1500 interspecific hybrid combinations in Abelmoschus in addition to over 200 cross combinations in Vigna and Cucumis.
- A user-friendly digitized system for identification and documentation of the species diversity in India was devised. This includes digitized images of all species studied and collected under this project. This database with distribution maps, plant images and descriptions along with the brief illustrated 'Fliers' for each of the genera is expected to be handy and useful to taxonomists and explorers as well as plant breeders for correct identification and description of species utilized in crop improvement programmes. This will reduce enormously the confusion prevailing in literature on species utilized in crossing programes.
Genomic studies in sesame germplasm for understanding the genetic basis of yield contributing traits in Sesame
Understanding the effects of domestication bottleneck and selection on fatty acid desaturases in Indian sesame germplasm:
Sesame (Sesamum indicum L.), is one of the oldest and most nutritional oilseed crop, whose domestication history has been poorly understood. The study conducted in sesame core germplasm suggests that sesame has undergone domestication bottleneck over a long period of time. In this analysis, we targeted 4.4 Mbp of the genomic DNA of sesame which comprised of stearoyl acyl desaturase (sad), fatty acid desaturase 2 (fad2) and omega 3 fatty acid desaturase (o3fad) genes in 99 selected accessions of four sesame germplasm populations groups, namely, wild, landrace, cultivar and introgressed. Results of this study showed that genetic diversity of the crop has been eroded due to selection after domestication as the cultivars and landraces lost 46.6% and 36.7% of nucleotide diversity, respectively. However, there was no significant reduction of genetic diversity in cultivars compared to landraces indicating that generation of improved cultivars through cross-breeding was in less frequency in this population. To evaluate the impact of selection across fatty acid biosynthetic pathway, we surveyed individual nucleotide diversity at three major genes. In our study, the analysis between wild and cultivars indicated positive selection in fad2 and o3fad loci. However, sad locus showed more diversity in cultivars compared to wild. Though locus-to-locus sequence variation was observed, positive results with two most important loci supported selection after domestication. Reduced diversity in these critical quality governing genes in cultivars suggests that future sesame breeding would benefit from the incorporation alleles from sesame’s wild relatives.
Molecular mapping and localization of yield related QTLs in sesame
Yield related QTLs were mapped on to the molecular maps generated using recombinant inbred lines (RILs) comprising 210 lines in F8 generation of a cross between a cultivated sesame and its closest progenitor species, Sesamum malabaricum. The traits scored were – days
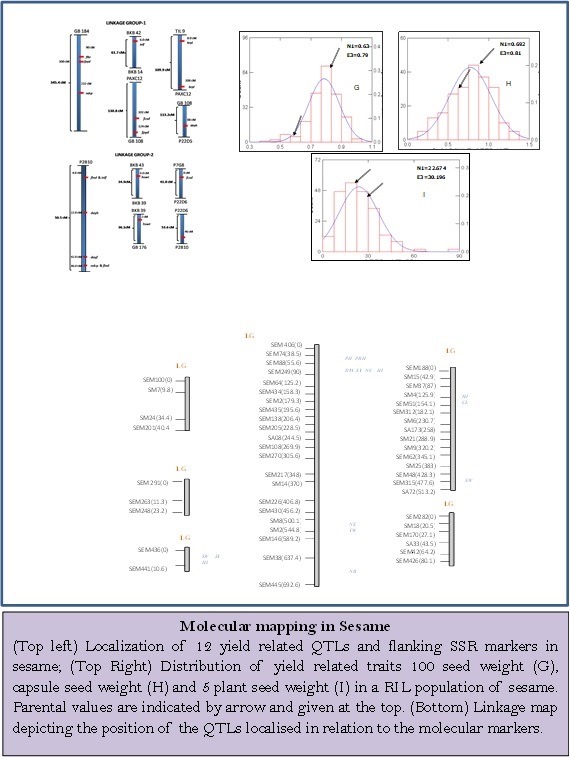
to flowering, days to 50% flowering, capsules per node, capsules per plant, hairiness of capsules, leaf hairiness, seed color, first capsule bearing node, leaf type, plant height, internode length for first five nodes, stem girth, capsule diameter, capsule length, days to maturity, 1000 seed weight, seed yield for 5 plants. Screening of 300 SSRs for parental polymorphism resulted in identification of 43 polymorphic SSR markers in the RIL population and 55 SSRs in Advance BC population. Finally, 33 SSRs showing no segregation distortion were placed on two linkage groups using RILs and for Advance BC population 55 markers were placed on six linkage groups. Review of these maps with the software JOINMAP resulted in a single linkage group comprising 22 markers over a length of 903.2 cM. Finally, a linkage map constructed with LOD score set to 5.0 and it included 23 markers in 3 linkage groups. The lengths of linkage groups were 466.4 cM, 416.3 cM and 99.5 cM respectively.
The interval mapping approach helped in localization of 12 QTLs for nine important traits, namely, number of branches per plant, date of 50% flowering, seed weight per capsule, seed yield per plant, internode length L1, stem girth, capsules per node, days to maturity and node with capsule. These QTLs were localized on to the two linkage groups identified in sesame. A frequency plot of the three important traits mapped is presented below which indicated normal distribution for these quantitative traits.
The linkage map depicts the position of the QTLs localised in relation to the molecular markers. The total linkage group size, relatie positions etc are expected to improve with greater saturation of the linkage groups. The linkage map constructed with advance BC population is presented in Fig which includes 55 SSR markers in 6 linkage groups. SSR markers and linkage maps with QTLs localized on them in such large numbers were not available in sesame till now. Availability of these genomic resources will provide greater and much needed impetus to crop improvement programmes in sesame which is fourth important oilseed crop with novel properties.
GM Detection
ICAR-NBPGR facilitates import of transgenics for research purpose (including their molecular testing) as per Government of India notification no. GSR 1067 (E) dated 5 Dec 1989, and Plant Quarantine (Regulation of Import into India) Order 2003 after technical clearance for import by RCGM. So far, >250 imported consignments of transgenics have been tested.
|
Rendering GMO Testing Services
|

|
|
As a Referral Laboratory to detect presence or absence of Genetically Modified Organisms (GMO) notified under sub-section (1) of Section 4 of the Seeds Act, 1966 in a Gazette of India Notification (Department of Agriculture and Farmers Welfare, DAFW) in 2017, the GM detection laboratory is rendering GMO testing services in the following areas:
- Suspected? imported seed consignments channeled through DAFW/ EXIM Committee for export and import of seeds for non-GMO confirmation
- Event confirmation in case of new Bt cotton hybrids
- Technical and scientific support to regulatory bodies
Resource generated (2018-2023): Approx Rs. 14.00 lakhs
|
GM Detection Technologies developed/ validated
- Validated PCR/Real-time PCR protocols available for GM detection in 13 Crops (Apple, Brinjal, Cotton, Flax, Indian mustard/Canola, Papaya, Pigeonpea, Maize, Rice, Soybean, Sweet pepper, Tomato, Wheat); Oil (Canola, Cottonseed, Mustard, Soybean); Food Products (bakery & confectionery, cereal products, baby food, snacks & instant mixes), in ISO17025 compliance.
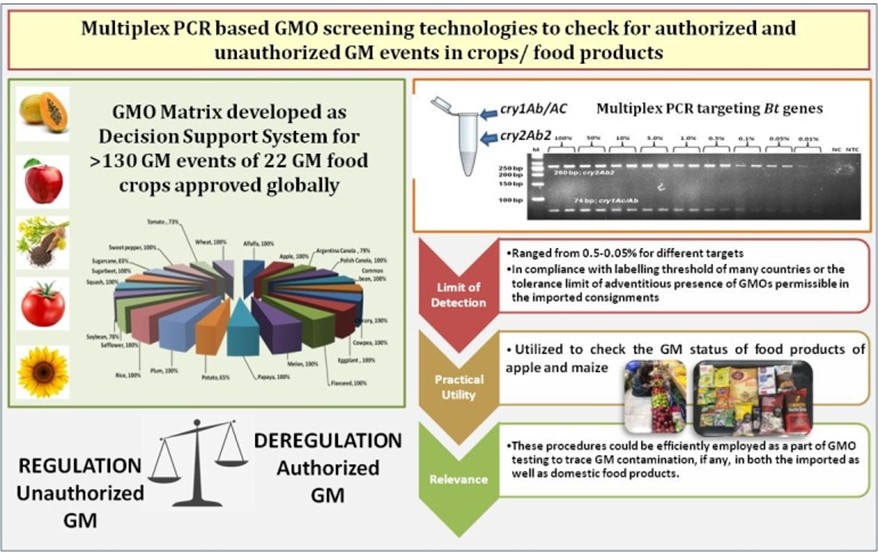
- GMO Matrix as Decision Support System
- Cost-effective GMO screening matrix developed for the first time in the country for detection of 141 GM events (commercialized/under field trials/ imported) (Food Cont 2014)
- Concept of crop-specific GMO matrix combined with multiplex PCR introduced to screen GM maize and cotton events approved globally (Food Cont 2016)
- GMO matrix compiled for 22 GM food crops based on which multiplex PCR-based GM diagnostics developed (J Plant Biochem Biotechnol 2023)
|
|
- Loop-mediated Isothermal Amplification (LAMP) technology for rapid/on-site GM detection
Visual/Real-time LAMP assays developed for >20 targets including promoters, markers genes, transgenes, construct regions and GM events (Eur Food Res Technol 2023, J AOAC Int, 2018, J Plant Biochem Biotechnol 2018, Food Control, 2015, 2017; 2019, J Agric Food Chem, 2013)
|
Promoters
|
P-35S, P-FMV, P-nos, P-ract
|
|
Marker genes
|
aadA, nptII, uidA, pat, pmi
|
|
Transgenes
|
cry1Ac, cry2Ab2, cp4-epsps
|
|
Constructs
|
P35S-cry1Ac, cry2Ab2-tnos, cp4epsps-tnos
|
|
Event specific
|
Bt cotton (MON531, MON15985);
GM maize (Bt11, GA21, MON810, MON89034, NK603, TC1507)
|
|
Terminator
|
pin II
|
 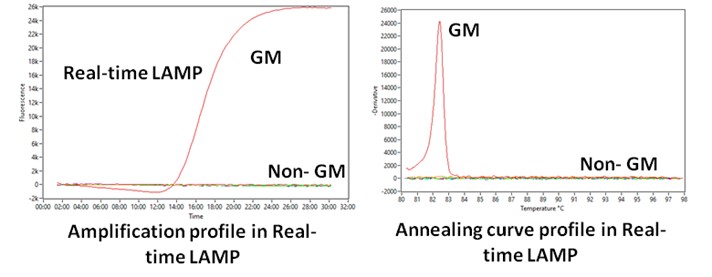
- Checking Unauthorized GMO in the National Genebank (NGB) and in Supply Chain
- For ensuring GM free conservation of germplasm, selected accessions of different crops conserved at National Genebank (as shown in the Figure) monitored for ensuring absence of adventitious presence of transgenes.
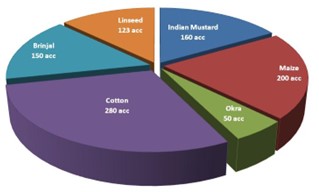
Cropwise number of accessions tested for GM status
- The unauthorized presence of GM brinjal checked in 100 samples collected from 14 villages of Maharashtra and in 211 brinjal samples collected from Assam, Meghalaya, Mizoram, Tripura, West Bengal. None of the samples showed adventitious presence of transgenes.
|
Commercialization of GM Detection Technologies
|
|
GM Detection Technologies transferred:
Five cost-efficient/rapid GMO screening technologies transferred to M/s DSS Imagetech Private Limited, New Delhi in August 2015
- Duplex real-time PCR for P-35S and T-nos
- Visual LAMP targeting eight targets
- Real-time LAMP targeting eight targets
- TaqMan® real-time PCR-based multi-target system covering 47 targets
- Hexaplex PCR targeting six marker genes
|
GMO Detection Kits:
Under BIRAC funded collaborative project, following GM detection kits commercialized by the DSS Imagetech Private Limited in 2022:
- GMO Detection Kit (35S/NOS): targeting CaMV 35S promoter and nos terminator
- FMV (GMO) Detection Kit: targeting FMV promoter
|
|
|
|
|
IPs and Technology Transfer
Patent No. 245749 : Process enabling simultaneous detection of two transgenes namely 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS or CP4EPSPS) gene and cauliflower
Inventors : Randhawa GJ, Firke PK and Karihaloo JL
Commercialization : Available on Non-exclusive basis
Patent No. 254341 : Process enabling simultaneous detection of two transgenes namely human serum albumin (HAS) and bar genes using a multiplex polymerase chain reaction
Inventors: Randhawa GJ, Firke PK and Karihaloo JL
Commercialization: Available on Non-exclusive basis
Patent No 258165: Diagnostic kit based on polymerase chain reaction for detection of cry1Ac gene
Inventors: Randhawa GJ and Firke PK
Commercialization: Available on Non-exclusive basis
Copyright Reg.No. 5760/06-CD (SW): Development of Crop DNA Fingerprinting Database Software Package
Author: Madhu Bala
Commercialization: Available on request for research use; for commercial use on non-exclusive basis
Copyright Reg. No. SW-15429/2022: AMARANTH GENOMIC RESOURCE DATABASE
Authors: Rakesh Singh, Akshay Singh, Ajay Kumar Mahato, S Rajkumar, A K Singh, Rakesh Bhardwaj, S K Kaushik, Sandeep Kumar and Veena Gupta
Commercialization: Freely Available for non-commercial purpose.
Copyright Reg. No. SW-15428/2022: TINOTRANSCRIPTDB (TINOSPORA CORDIFOLIA TRANSCRIPTS & SSR DATABASE)
Authors: Rakesh Singh, Akshay Singh, Ajay Kumar Mahato, Rajesh Kumar, Amit K. Singh, Sundeep Kumar, Soma S. Marla, Ashok Kumar and Nagendra K. Singh
Commercialization: Freely Available for non-commercial purpose.
Copyright Reg. No. SW-15900/2022: APTRANSDB: ANDROGRAPHIS TRANSCRIPTS & SSR DATABASE
Authors: Rakesh Singh, Akshay Singh, Ajay Kumar Mahato, Rajesh Kumar, Amit K. Singh, Sundeep Kumar, Soma S. Marla, Ashok Kumar, and NK Singh
Commercialization: Freely Available for non-commercial purpose.
|
|
|
-
Dr. Dhammaprakash Pandhari Wankhede received the best poster award in the 1st National Conference on Plant Genetic Resources Management, November 22-24, 2022 organized at NASC, New Delhi by the Indian Society of Plant Genetic Resources, New Delhi, India.
-
Dr. Dhammaprakash Pandhari Wankhede received the Best Paper Award for oral presentation: in the ISPP North Zonal Seminar-2022 on Inter-Disciplinary Research Strategies for Climate Resilient Agriculture held at ICAR-Sugarcane Breeding Institute, Regional Centre, Karnal, on 25th June 2022
-
Dr Monika Singh received “Young Women Scientist Award (Agricultural Biotechnology)” by Royal Association for Science-Led Socio-Cultural Advancement (RASSA) and Sardarkrushinagar Dantiwada Agricultural University (SDAU) in National Conference on Natural Farming for Sustainable Agriculture and National Prosperity (11-13 Nov 2022) at SDAU, Palanpur
-
Dr. Dhammaprakash Pandhari Wankhede received the Early Career Research Award (ECRA) in the form research project for three years (2018-2021) by SCIENCE & ENGINEERING RESEARCH BOARD (SERB), Department of Science and Technology, Government of India
-
Dr Monika Singh received “Kanwar Virender Singh Memorial All India Best Publication Award 2021 - By SADHNA (Society for Advancement of Human and Nature), Dr YS Parmar University of Horticulture and Forestry, Nauni, Solan (Himachal Pradesh) Efficient DNA extraction procedures for processed food derivatives: a critical step to ensure quality for GMO analysis. Food Analytical Methods 14: 2249-2261
-
Dr Gurinderjit Randhawa received “Dr Panjabrao Deshmukh Outstanding Women Scientist Award 2020” by the Indian Council of Agricultural Research, New Delhi.
-
Dr. Gurinderjit Randhawa has been elected “NAAS Fellow 2020” by National Academy of Agricultural Sciences, New Delhi.
-
Dr. Yasin Jeshima K. conferred “Fellow of Young Academy of India 2021” by Young Academy of India.
-
Dr Rakesh Singh received "Dr B R Barwale Award" for Application/Excellence in Plant Genetic Resources by Indian Society of Plant Genetic Resources (ISPGR), New Delhi for the year 2020.
-
Dr Amit Kumar Singh conferred Fellow of Indian Society of Genetics and Plant Breeding (ISGPB), New Delhi for the year 2020.
-
Dr Sanjeev Kumar Singh conferred “Yashsvi Samman” for the distinguished services in the field of Science and Society by Royal Association for Science-led-socio-cultural Advancement (RASSA), New Delhi for the year 2020.
-
Dr. Parimalan R. received “Dr. APJ Abdul Kalam Best Scientist Award” for the year 2020 by Bose Science Society, Pudukkotai, Tamilnadu.
-
Dr. Yasin Jeshima K. conferred “Outstanding International Researcher Award” by Scientific Society for International Institute of Organized Research for the year 2020-21.
-
Dr R Parimalan conferred Fellow of Indian Society of Plant Genetics Resources (ISPGR), New Delhi for the year 2019.
-
Dr. Sanjeev Kumar Singh received “Best Oral Presentation Award” for the paper “Characterization of Rice bean [Vigna umbellata (Thumb.) Ohwi & Ohashi] Landraces from Northeast India” in “International Seminar on Sustainable Agricultural Development in Changing Global Scenario at Banaras Hindu University, Varanasi from 11th to 13th Oct. 2019.
- Dr. Amit Kumar Singh received “Dr. R S Paroda Young Scientist Award” for the year 2018 by ISPGR Society New Delhi.
- Dr. Sundeep Kumar conferred Fellow of Indian Society of Genetics and Plant Breeding, New Delhi for the year 2018.
- Dr. Rakesh Singh received "RK Arora Best Research Paper Award" for the year 2017 by Indian Society of Plant Genetic Resources, New Delhi.
- Dr. R. Parimalan received Michael D Gale travel grant award for the best abstract under plant category in 25thPlant and Animal Genome Conference, January 14-18, 2017, San Diego, USA.
- Dr. R. Parimalan received "Dr. RS Paroda Young Scientist Award" for significant contributions in the field of Plant Genetic Resources for the year 2017.
- Dr. R. Parimalan selected Honorary Faculty at Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Australia.
- Dr. Sanjeev Kumar Singh received "Best Oral Presentation Award" in the National seminar on Smart Farming for Enhancing Input Use Efficiency, Income and Environmental Security, at ICAR Research Complex for NEH Region, Umiam, Meghalaya, September 19-21, 2017.
- Dr. Monika Singh received "Young Scientist Award" at the 3rd International Conference on Bioresource and Stress management at State Institute of Agriculture Management, Jaipur, November 8-11, 2017.
- Dr. Sundeep Kumar received "Distinguished Scientist Award" in International Conference on Global Research Initiatives for Sustainable Agriculture & Allied Sciences (GRISAAS-2017) at Maharana Pratap University of Agriculture & Technology, Udaipur Rajasthan, December 02-04, 2017.
- Dr. G J Randhawa received "Agriculture Leadership Award 2015" by Agriculture Today for leading role in Biotechnology Research and Development during 2015.
- Dr. G J Randhawa received "Best Scientist Award" by Science and Technology, EET CRS, Research Wing for excellence in Professional Education and Industry, New Delhi during 2015.
- Dr. G J Randhawa received the recognition by OMICS International, USA for outstanding contributions in 7th Indo Global Summit and Expo on Food and Beverage during 2015.
- Dr. Rakesh Singh conferred Fellow of Indian Society of Genetics and Plant Breeding, New Delhi for the year 2015.
- Dr. Rakesh Singh conferred Fellow of Indian Society of Plant Genetic Resources, New Delhi for the year 2012.
- Dr. Rakesh Singh received "Best Research Paper Award" by the Horticultural Society of India, IARI, New Delhi for the year 2011.
- Dr. Rakesh Singh received "Young Scientist Award (Biotechnology)" for significant contributions in the field of Plant Biotechnology by the Society for Plant Research, S.V.P. University of Agriculture and Technology, Meerut during 2010.
- Dr. Rakesh Singh received "Dr RS Paroda Young Scientist Award" for significant contributions in the field of Plant Genetic Resources, 2007-2008.
Awards and recognitions
Shri Narendra Singh Tomar, Hon’ble Union Minister of Agriculture and Farmers Welfare, Govt. of India handing over the certificate of ISO/IEC 17025:2017 Accreditation of GM Detection Research Facility to Dr Gurinderjit Randhawa, Head, Division of Genomic Resources, ICAR-NBPGR, New Delhi on 16th August, 2021.

Dr Gurinderjit Randhawa, Head & Principal Scientist, Division of Genomic Resources, ICAR-NBPGR, New Delhi received “Dr Panjabrao Deshmukh Outstanding Women Scientist Award 2020” by the Indian Council of Agricultural Research on dated 16th July, 2021.

Agriculture Leadership Award: Dr. Gurinderjit Randhawa received Agriculture Leadership Award 2015 for leading role in Biotechnology Research and Development by Agriculture Today on 18 September, 2015 at New Delhi.
Krishi Jeevan Jyoti Award: Dr. Mahesh C Yadav, Principal Scientist, ICAR-NBPGR, New Delhi was conferred “Krishi Jeevan Jyoti Award 2015” on 27-01-2016 at NASC Complex, New Delhi for his outstanding contribution in research in the field of Genetics and training of youth in Agricultural Biotechnology.
|
|
|
- Molecular Profiling and DNA fingerprinting of released varieties of select crops
- Molecular diagnostics for GM detection
- Genomic Resource Generation and Bioinformatics
|
|
|
|
|
|
1.
|
Choudhury DR, R Kumar, A Maurya, DP Semwal, RS Rathi, RK Gautam, AK Trivedi, SK Bishnoi, SP Ahlawat, Kuldeep Singh, NK Singh, and R Singh (2023). SSR and SNP marker-based investigation of Indian rice land-races in relation to their genetic diversity, population structure, and geographical isolation. Agriculture, 13(4): 823
|
|
|
|
|
|
|
|
|
|
|
6.
|
Ruperao P, P Bajaj, R Subramani, R Yadav, VBR Lachagari, SP Lekkala, A Rathore, S Archak, UB Angadi, R Singh, K Singh, S Mayes, and R Parimalan (2023). A pilot-scale comparison between single and double-digest RAD markers generated using GBS strategy in sesame (Sesamum indicum L.). PLoS One, 18: e0286599:
|
|
|
|
|
|
|
|
|
|
|
|
|
12.
|
Singh A, AK Mahato, A Maurya, R Subramani, AK Singh, R Bhardwaj, SK Kaushik, S Kumar, V Gupta, K Singh and R Singh (2023). Amaranth Genomic Resource Database (AGRDB): an integrated database resource of Amaranth genes and genomics. Frontiers in Plant Science, 14:1203855:
|
|
|
|
|
14.
|
Srivastav M, N Radadiya, S Ramachandra, P Jayswal, N Singh, S Singh, AK Mahato, G Tandon, A Gupta, R Devi, SH Subrayagowda, G Kumar, P Prakash, S Singh, N Sharma, A. Nagaraja, A Kar, SG Rudra, S Sethi, S Jaiswal, MA Iquebal, R Singh, SK Singh and NK Singh (2023). High resolution mapping of QTLs for fruit color and firmness in Amrapali/Sensation mango hybrids. Frontiers in Plant Science, 14-1135285:
|
|
|
15.
|
Vats G, D Das, R Gupta, A Singh, A Maurya, S. Rajkumar, AK Singh, R Bhardwaj, S Kumar, SK Kaushik, V Gupta, K Singh and R Singh (2023). Validation of Genome-wide SSR markers developed for genetic diversity and population structure study in Grain Amaranth (Amaranthus hypochondriacus). Agriculture, 13(2): 431
|
|
|
|
|
17.
|
Ankit Saroha, Sunil S Gomashe, VikenderKaur, Deepa Pal, ShraddhaUjjainwal, J Aravind, Mamta Singh, S Rajkumar, Kuldeep Singh, Ashok Kumar, Dhammaprakash Pandhari Wankhede (2023). Genetic dissection of thousand-seed weight in linseed (Linumusitatissimum L.) using multi-locus genome-wide association study.. Frontiers in Plant Science, 14, 1166728:
|
|
|
18.
|
Kumar, G.P.; Pathania, P.; Goyal, N.; Gupta, N.; Parimalan, R.; Radhamani, J.; Gomashe, S.S.; Kadirvel, P.; Rajkumar, S. (2023). Genetic Diversity and Population Structure Analysis to Construct a Core Collection from Safflower (Carthamustinctorius L.) Germplasm through SSR Markers.. Agriculture, 13: 836
|
|
|
|
|
20.
|
Bansal, S., Sharma, M.K., Singh, S. Joshi, P., Pathania, P., Malhotra, EV., S. Rajkumar, Misra, P. (2023). Histological and molecular insights in to in vitro regeneration pattern of Xanthosomasagittifolium.. Scientific Reports, 13(1): 5806
|
|
|
21.
|
Francis, A., Singh, N.P., Singh, M., Sharma, P., Gayacharan, Kumar, D., Basu, U., Bajaj, D., Varshney, N., Joshi, D.C., Semwal, D.P., Tyagi, V., Wankhede, D., Bharadwaj, R., Singh, A.K., Parida, S.K. and Chattopadhyay, D. (2023). The ricebean genome provides insight into Vigna genome evolution and facilitates genetic enhancement.. Plant Biotechnol. J.,, 21(8): 1522
|
|
|
22.
|
Kaur V, Singh M, Wankhede DP, Gupta K, Langyan S, Aravind J, Thangavel B, Yadav SK, Kalia S, Singh K and Kumar A (2023). Diversity of Linum genetic resources in global genebanks: from agro-morphological characterisation to novel genomic technologies – a review. Frontiers in Nutrition, 10:1165580:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1.
|
T Danakumara, J Kumari, AK Singh, SK Sinha, AK Pradhan, S Sharma, SK Jha, R Bansal, S Kumar, GK Jha, MC Yadav and PVV Prasad (2022). Genetic dissection of seedling root system architectural traits in a diverse panel of hexaploid wheat through multi-locus genome-wide association mapping for improving drought tolerance. Int. J. Mol. Sci., 22(13):7188:
|
|
|
2.
|
Tripathi K, J Kumari, PG Gore, DC Mishra, AK Singh, GP Mishra, C Gayacharan, HK Dikshit, N Singh, DP Semwal, R Mehra, R Bhardwaj, R Bansal, JC Rana, A Kumar, V Gupta, K Singh and A Sarker (2022). Agro-morphological characterization of lentil germplasm of Indian National Genebank and development of a core set for efficient utilization in lentil improvement programs. Front. Plant Sci., 12:751429:
|
|
|
|
|
4.
|
Saroha A, D Pal, V Kaur, S Kumar, A Bartwal, Aravind J, Radhamani J, Rajkumar S, R Kumar, SS Gomashe, A Sengupta, DP Wankhede (2022). Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time.. Genet Resour Crop Evo, 69: 315-333
|
|
|
5.
|
Gangappa ND, C Singh, MK Verma, M Thakre, AMV Sevanthi, R Singh, M Srivastav, Raghunandan K, C Anusha, V Yadav, A Nagaraja (2022). Assessing the genetic diversity of guava germplasm characterized by morpho-biochemical traits. Frontiers in Nutrition, 9:1017680:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
15.
|
Singh R, AK Mahato, A Singh, R Kumar, A K Singh, S Kumar, SS Marla, A Kumar, NK Singh (2022). TinoTranscriptDB: Transcripts and Microsatellite Markers Database of Tinospora cordifolia, an Important Medicinal Plant. Genes, 13: 14 33
|
|
|
|
|
|
|
|
|
|
|
|
|
21.
|
Padhi SR, John R, Bartwal A, Tripathi K, Gupta K, Wankhede DP, Mishra GP, Kumar S, Rana JC, Riar A and Bhardwaj R (2022). Development and optimization of NIRS prediction models for simultaneous multi-trait assessment in diverse cowpea germplasm.. Frontiers in Nutrition, 9:1001551:
|
|
|
|
|
|
|
24.
|
Panigrahi, S. K., Tripathi, K., Singh, R., Kumar, R., Sanghamitra, P., Wankhede, D. P., Singh, N., Dubey, K. K. D., & Gupta, K. (2022). Evaluation of black gram (Vignamungo) genepool against Callosobruchusmaculatus and diversity analysis inter se.. The Indian Journal of Agricultural Sciences, 92(7), 915–919:
|
|
|
|
|
26.
|
Saroha A, Pal D, Gomashe SS, Akash, Kaur V, Ujjainwal S, Rajkumar S, Aravind J, Radhamani J, Kumar R, Chand D, Sengupta A and Wankhede DP* (2022). Identification of QTNs Associated with Flowering Time, Maturity, and Plant Height Traits in Linum usitatissimum L. Using Genome-Wide Association Study. . Frontiers in Genetics, 13:811924:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4.
|
Verma SK, S Mittal, Gayacharan, DP Wankhede, SK Parida, D Chattopadhyay, G Prasad, DC Mishra, DC Joshi, M Singh, K Singh and AK Singh (2021). Transcriptome analysis reveals key pathways and candidate genes controlling seed development and size in rice bean (Vigna umbellata). Frontiers in Genetics, 12:791355:
|
|
|
5.
|
Mishra GP, MS Aski, T Bosamia, S Chaurasia, DC Mishra, J Bhati, A Kumar, S Javeria, K Tripathi, M Kohli, RR Kumar, AK Singh, J Devi, S Kumar and HK Dikshit (2021). Insights into the host-pathogen interaction pathways through rna-seq analysis of Lens culinaris Medik. in Response to Rhizoctonia bataticola Infection. Genes, 13(1):90:
|
|
|
6.
|
Priya S, R Bansal, G Kumar, HK Dikshit, J Kumari, R Pandey, AK Singh, K Tripathi, N Singh, NKP Kumari, S Kumar and A Kumar. (2021). Root trait variation in lentil (Lens culinaris Medikus) germplasm under drought stress. Plants, 10(11):2410:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
13.
|
?Srivastav M, SK Singh, J Prakash, R Singh, N Sharma, S Ramchandra, R Devi, A Gupta, AK Mahto, PK. Jayaswal, S Singh and NK Singh (2021). New hyper-variable SSRs for diversity analysis in mango (Mangifera indica L.).. Indian J. Genet., 81(1): 119-126.
|
|
|
|
|
|
|
|
|
17.
|
Gowthami, R., Sharma, N., Gangopadhyay, K. K., Rajkumar, S., Pathania, P., & Agrawal, A (2021). Cryopreservation of Pollen of Abelmoschus Moschatus Medik. Subsp. Moschatus as an Aid to Overcome Asynchronous Flowering for Wide Hybridization with Cultivated Okra [A. Esculentus (l.) Moench]. Cryoletters, 42(4): 233-244
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6.
|
Singh S, Mahato AK, Jayaswal PK, Singh N, Dheer M, Goel P, Raje RS, Yasin JK, Sreevathsa R, Rai V, Gaikwad K. Singh N K. (2020). A 62K genic-SNP chip array for genetic studies and breeding applications in pigeonpea (Cajanus cajan L. Millsp.).. Scientific reports, 10(1): 1-4.
|
|
|
|
|
|
|
9.
|
Rani K, BR Raghu, SK Jha, P Agarwal, N Mallick, M Niranjana, JB Sharma, AK Singh, NK Sharma, SMS Tomar and Vinod (2020). A novel leaf rust resistance gene introgressed from Aegilops markgrafii maps on chromosome 2AS.. Theor. and App. Gen., 56:: 126
|
|
|
|
|
11.
|
Singh A, Y Singh, A Mahato, P Jayaswal, S Singh, R Singh, N Yadav, A Singh, P Singh, R Singh, R Kumar, E Septiningsih, H Balyan, N K Singh (2020). Allelic sequence variation in the Sub1A, Sub1B and Sub1C genes among diverse rice cultivars and its association with submergence tolerance.. Scientific Reports., 10:: 8621
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7.
|
Kumari R, DP Wankhede, A Bajpai, A Maurya, K Prasad, D Gautam, P Rangan, M Latha, KJ John, A Suma, KV Bhat and AB Gaikwad (2019). Genome wide identification and characterization of microsatellite markers in black pepper (Piper nigrum): A valuable resource for boosting genomics applications.. PLoS One., 13:: 14(12): e0226002. doi:10371/journal.pone.0226002.
|
|
|
8.
|
Kumar S, BS Phogat, Vikas VK, AK Sharma, MS Saharan, AK Singh, J Kumari, R Singh, SR Jacob, GP Singh, M Sivasamy, P Jayaprakash, Madhu Meeta, JP Jaiswal, Deep Shikha, BK Honrao, IK Kalappanavar, PC Mishra, SP Singh, SS Vaish and VA Solanki (2019). Mining of Indian wheat germplasm collection for adult plant resistance to leaf rust.. PLoS ONE., 14(3):: e0213468.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2.
|
Singh, S K, P C Kole, A K Misra, Somnath Roy, Lalit Arya, Manjusha Verma, R. Bhardwaj, P. Suneja, Med Ram Verma, KV Bhat and Rakesh Singh (2017). Characterization of Perilla frutescens (Linn.) Britt based on morphological, biochemical and STMS markers
. Industrial Crops & Products, 109: 773–785
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
22.
|
Singh S K, PC Kole, AK Misra, S Roy, L Arya, M Verma, R Bhardwaj, P Suneja, MR Verma, KV Bhat and R Singh* (2017). Characterization of Perilla frutescens (Linn.) Britt based on morphological, biochemical and STMS markers.. Industrial Crops and Products, 109C: 773-785
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5.
|
Archak S, Tyagi RK, Harer PN, Mahase LB, Singh N, Om P Dahiya, Nizar MA, Singh M, Tilekar V, Kumar V, Dutta M, Singh NP and Bansal KC (2016). Characterization of chickpea germplasm conserved in the Indian National Genebank and development of a core set using qualitative and quantitative trait data. The Crop Journal, 4: 1-8
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7.
|
Roy, S, A Banerjee, B Mawkhlieng, A K Misra, A Pattanayak, G D Harish, S K Singh, S V Ngachan and K C Bansal (2015). Genetic diversity and population structure in aromatic and quality rice (Oryza sativa L.) landraces from North-Eastern India.. PLoS ONE, 10(6), e0129607.:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9.
|
V. K. Vikas, M. Sivasamy, J. Kumar, P. Jayaprakash, S Kumar, R. parimalan, A Kumar, K Srinivasan, J. Radhamani, S R Jacob, M Yadav, J Rani, I S Bist, D C Bhandari, S Archak, M Datta R K Tyagi and K C Bansal (2014). Stem and leaf rust resistance in wild relatives of wheat with D genome. Genetic Resources and Crop Evolution, 61: 861-874
|
|
|
|
|
11.
|
Vikas VK, Sivasamy M, Kumar J, Jayaprakash P, Kumar S, Parimalan R, Kumar A, Srinivasan K, Radhamani J, Jacob S, Yadav M, Rani J, Bisht IS, Bhandari DC, Archak S, Dutta M, Tyagi RK and Bansal KC (2014). Stem and leaf rust resistance in wild relatives of wheat with D genome (Aegilops spp.). Genetic Resources and Crop Evolution, 61: 861-874
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
17.
|
Singh SK, GR Lavanya, KV Bhat, GS Babu, L. Arya, M Verma, Z Hussain, S Roy, RS Rathi, AK Misra (2012). Microsatellite markers revealed genetic diversity in mung bean mutant lines.. Indian J. Hill Farm., 25(1): 38-43.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Inhouse Projects
|
| Programme | Project Title | Leader and Associates | Project Code |
|
1
|
Development of genomic tools for identification, protection and enhanced utilization of PGRs
|
Gurinderjit Randhawa
|
PGR/DGR-BUR-DEL-01.00
|
|
1.1
|
Development of genomic tools for discovery and validation of genes of economic importance for enhancing the use of plant genetic resources of pulses and oilseed crops
|
Rajesh Kumar, S Marla, J Radhamani, Yasin JK, DP Wankhede, Rashmi Yadav, S Rajkumar, R Parimalan, Amit Kumar Singh, Vikender Kaur and SK Singh
|
PGR/DGR-BUR-DEL-01.01
|
|
1.2
|
Development of genomic tools for enhanced utilization of cereals
|
Rakesh Singh, MC Yadav, Sundeep Kumar, AK Singh, R Parimalan and Sheel Yadav
|
PGR/DGR-BUR-DEL-01.02
|
|
1.3
|
Development of genomic tools for enhanced utilization of millets
|
Lalit Arya, Monika Singh, Mamta Singh, Sunil Gomashe and Shashi Bhushan Choudhary
|
PGR/DGR-BUR-DEL-01.03
|
|
1.4
|
Development of genomic tools for enhanced utilization of horticultural crops
|
AB Gaikwad, M Verma, S Archak, K Joseph John and Dikshant Gautam
|
PGR/DGR-BUR-DEL-01.04
|
|
1.5
|
Development of genomic tools for species delineation and genetic erosion studies in selected crops.
|
MC Yadav, S Rajkumar, S Marla, J Radha Mani, DR Pani and M Latha
|
PGR/DGR-BUR-DEL-01.05
|
|
1.6
|
Establishment and maintenance of national genomic resources repository and bioinformatics facility
|
Sundeep Kumar, MC Yadav, MK Rana, Lalit Arya, M Verma, S Raj Kumar, Rajesh Kumar, AK Singh, JK Yasin, R Parimalan, Sheel Yadav (on study leave w.e.f. 28 July 2018), DP Wankhede, Monika Singh, Sangita Bansal and SK Singh
|
PGR/DGR-BUR-DEL-01.06
|
|
1.7
|
Development and utilization of GM diagnostics for detection of genetically engineered plants and derivatives
|
Monika Singh, Gurinderjit Randhawa
|
PGR/DGR-BUR-DEL-01.07
|
|
1.8
|
Development of unique identity system for cultivars and genetic stocks for IPR protection
|
MK Rana, AB Gaikwad, Rakesh Singh, Lalit Arya, Manjusha Verma, Sundeep Kumar, Rajesh Kumar, S Rajkumar, R Parimalan, AK Singh, Sheel Yadav (on study leave w.e.f. 28 July 2018), DP Wankhede, Yasin JK and SK Singh
|
PGR/DGR-BUR-DEL-01.08
|
|
|
|
Externally Funded Projects
|
| Programme | Project Title | Funding Agency | Principle Investigator | Date of Start | Date of Termination | Budget (Lakhs) | Project Code |
|
001
|
Rationalisation of rice collections originating from major areas of diversity and allele mining in selected unique set of accessions for biotic, abiotic and quality traits using molecular markers.
|
ICAR
|
Rakesh Singh
|
April 2021
|
March 2026
|
339.58
|
1001381
|
|
002
|
National Programme for Quarantine and GM Diagnostics of Genetically Engineered Plant Material (Component-2)
|
DBT
|
Monika Singh
|
September 2020
|
September 2025
|
278.56
|
1012550
|
|
003
|
Identification and validation of climate-smart germplasm and pre-breeding for abiotic stress tolerance in wheat and rice using crop wild relatives and related species
|
ICAR
|
Mahesh Chandra Yadav
|
April 2021
|
March 2026
|
534.00
|
OXX04063-ICAR-MY-
|
|
004
|
Genomics of black pepper, cardamom and fenugreek for relating genes to traits of economic importance
|
ICAR
|
Ambika Gaikwad
|
October 2017
|
March 2021
|
139.75
|
1611316024
|
|
006
|
Integrated genomics strategy for accelerating domestication of rice bean (Vigna umbellata)
|
DBT
|
Amit Kumar Singh
|
October 2018
|
October 2023
|
103.90
|
1010669
|
|
007
|
Characterization, evaluation of genetic resources for genetic enhancement and improvement of minor pulses
|
DBT
|
DP Wankhede
|
October 2018
|
October 2023
|
530.84
|
Pulses-I/2017-18
|
|
008
|
Development of amaranth core collection using SSR and SNP markers and evaluation of core set for nutritional, yield traits and abiotic stress tolerance
|
DBT
|
Rakesh Singh
|
March 2019
|
March 2024
|
212.54
|
1011049
|
|
010
|
Development of reference genome and core set using molecular markers: under network project 'Leveraging genetic resources for accelerated genetic improvement of linseed using comprehensive genomics and phenotyping approaches'
|
DBT
|
DP Wankhede
|
February 2020
|
February 2025
|
305.95
|
Linseed/2019-20
|
|
011
|
Genotyping of sesame germplasm following high throughput genomics
|
DBT
|
Parimalan R
|
February 2020
|
February 2025
|
1000.00
|
16113200037
|
|
012
|
Germplasm characterization and trait discovery in wheat using genomics approaches and its integration for improving climate resilience, productivity and nutritional
|
DBT
|
Sundeep Kumar
|
February 2020
|
February 2025
|
127.12
|
1012159
|
|
013
|
Germplasm genomics for trait discovery in wheat
|
DBT
|
Amit Kumar Singh & Sundeep Kumar
|
February 2020
|
February 2025
|
862.00
|
1012152
|
|
014
|
Capacity building in the area of genomic selection and high throughput phenotyping and its integration in national breeding programs
|
DBT
|
Sundeep Kumar
|
February 2020
|
February 2025
|
48.00
|
1012158
|
|
015
|
Evaluation of wheat germplasm for quality traits
|
DBT
|
Sundeep Kumar
|
February 2020
|
February 2025
|
51.36
|
1012156
|
|
016
|
Exploiting genetic diversity for improvement of safflower through genomics-assisted discovery of QTLs/Genes associated with agronomic traits
|
DBT
|
S Rajkumar
|
February 2020
|
February 2025
|
597.88
|
1012259
|
|
017
|
Mainstreaming rice landraces diversity in varietal development through genome- wide association studies: A model for large-scale utilization of gene bank collections of rice
|
DBT
|
Rakesh Singh
|
March 2020
|
March 2025
|
2227.90
|
1012179
|
|
018
|
Development of superior haplotype based near isogenic lines (Haplo-NILs) for enhanced genetic gain in rice
|
DBT
|
Rakesh Singh
|
March 2020
|
March 2024
|
157.12
|
1012181
|
|
019
|
Towards understanding the C3-C4 intermediate pathway in Poaceae and functionality of C4 genes in rice
|
ICAR
|
Parimalan R
|
April 2021
|
March 2026
|
150.00
|
1006976
|
|
020
|
Identification of genes/genomic regions associated with Fusarium head blight resistance in wheat
|
ICAR
|
Sundeep Kumar
|
April 2021
|
March 2026
|
85.00
|
1004936
|
|
021
|
A National Mission Mode Program on Nutritional improvement of digestible protein content and quality in rice.
|
DBT
|
Rakesh Singh
|
January 2022
|
January 2027
|
101.22
|
1013419
|
|
|
|
|
|
|
|
|
|
Dr. Rakesh Singh, Head of Division
|
|
Division of Genomic Resources
|
|
Phone: +91 9818934006, 011-25802884
|
|
Email: rakesh.singh2(AT)icar.gov.in, singhnbpgr(AT)yahoo.com
|
|
|
|
|
|
|
|
Dr. Lalit Arya, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: 011-25802797
|
|
Email: Lalit.Arya(AT)icar.gov.in
|
|
|
|
|
|
|
|
Dr. Sangita Bansal, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: Sangita.Bansal(AT)icar.gov.in, sangitabansal(AT)yahoo.com
|
|
|
|
|
|
|
|
Dr. Ambika Baldev Gaikwad, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: +91 9968277849
|
|
Email: Ambika.Gaikwad(AT)icar.gov.in
|
|
|
|
|
|
|
|
Dr. Sundeep Kumar, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: 9013575627
|
|
Email: Sundeep.Kumar(AT)icar.gov.in, sundeep(AT)daad-alumni.de
|
|
|
|
|
|
|
|
Dr. Rajesh Kumar, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: Rajesh.Kumar13(AT)icar.gov.in, kraj.pgr(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Vishnu Kumar, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: 011-25802810
|
|
Email: vishnu.kumar(AT)icar.gov.in
|
|
|
|
|
|
|
|
Dr. Parimalan R, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: +91 9968599479
|
|
Email: r.parimalan(AT)icar.gov.in, parimalan.r(AT)gmail.com, p.rangan(AT)uq.edu.au
|
|
|
|
|
|
|
|
Dr. S. Rajkumar, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: s.rajkumar(AT)icar.gov.in, rajnbpgr(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Mukesh Kr. Rana, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: 011-25802809 (Ext.809)
|
|
Email: Mukesh.Rana(AT)icar.gov.in
|
|
|
|
|
|
|
|
Dr. Manjusha Verma, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: Manjusha.Verma(AT)icar.gov.in, manjusha_v(AT)yahoo.com
|
|
|
|
|
|
|
|
Dr. Mahesh C. Yadav, Principal Scientist
|
|
Division of Genomic Resources
|
|
Phone: 011-25843697, 25841022 Ext.282; Mob +91 8278716712
|
|
Email: Mahesh.Yadav1(AT)icar.gov.in, mcyadav(AT)yahoo.com, maheshgenome(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Yasin Jeshima K, Senior Scientist
|
|
Division of Genomic Resources
|
|
Phone: +91 9968614672
|
|
Email: Yasin.Jeshima(AT)icar.gov.in, yasinlab2.icar(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Amit Kumar Singh, Senior Scientist
|
|
Division of Genomic Resources
|
|
Phone: +91 9968449805
|
|
Email: Amit.Singh5(AT)icar.gov.in, amit_singh79(AT)yahoo.com
|
|
|
|
|
|
|
|
Dr. Monika Singh, Senior Scientist
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: monika.singh(AT)icar.gov.in, monikasinghnbpgr(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Dhammaprakash Wankhede, Senior Scientist
|
|
Division of Genomic Resources
|
|
Phone: +91 9868028587
|
|
Email: d.wankhede(AT)icar.gov.in, wdhammaprakash(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Gayacharan , Scientist SS
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: gayacharan(AT)icar.gov.in
|
|
|
|
|
|
|
|
Dr. Laxmi Sharma, Scientist SS
|
|
Division of Genomic Resources
|
|
Phone: +91 8191001032
|
|
Email: laxmi.sharma(AT)icar.gov.in, laxmi1408(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Sheel Yadav, Scientist SS
|
|
Division of Genomic Resources
|
|
Phone: +91 9711187413
|
|
Email: Sheel.Yadav(AT)icar.gov.in, sheel.y85(AT)gmail.com
|
|
|
|
|
|
|
|
Dr. Madhu Bala, Scientist
|
|
Division of Genomic Resources
|
|
Phone: +91 9968077738
|
|
Email: M.Priyadarshi(AT)icar.gov.in, madhu74_nbpgr(AT)yahoo.com
|
|
|
|
|
|
|
|
|
|
Mr. Dikshant Gautam, Assistant Chief Technical Officer
|
|
Division of Genomic Resources
|
|
Phone: 8447216980
|
|
Email: Dikshant.Gautam(AT)icar.gov.in, dikshant7874(AT)hotmail.com, dikshant7874(AT)rediffmail.com
|
|
|
|
|
|
|
|
Mr. Manish Tomar, Technical Officer
|
|
Division of Genomic Resources
|
|
Phone: +91-9885222613
|
|
Email: manish.tomar(AT)icar.gov.in, manishtomar028(AT)gmail.com
|
|
|
|
|
|
|
|
Mrs. Akansha Bajpai, Technical Assistant
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: Akansha.Bajpai(AT)icar.gov.in, akanshabajpai11(AT)gmail.com
|
|
|
|
|
|
|
|
Ms. Kushaldeep Kaur Sodhi, Technical Assistant
|
|
Division of Genomic Resources
|
|
Phone: 7703999187
|
|
Email: Kushaldeep.KaurSodhi(AT)icar.gov.in, kushaldeepkaursodhi(AT)gmail.com
|
|
|
|
|
|
|
|
Mr. Ramesh Chand, Technician
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email: Ramesh.Chand7(AT)icar.gov.in
|
|
|
|
|
|
|
|
Mrs. Aggya Devi, Skilled Supporting Staff SSS1
|
|
Division of Genomic Resources
|
|
Phone:
|
|
Email:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|